3CR Bioscience reduces loss of lambs through rapid ID of genetic markers
49 percent of lamb mortality occurs within the first 48 hours following birth*, which is devastating for farmers and economically impactful. A new tool, developed by 3CR Bioscience, is making it easier for breeders to detect recessive gene variations that can be lethal when present in both parents.
Differences between individuals of the same species are known as traits and may result from particular sequences in the animals’ DNA. New tools developed by 3CR Bioscience for genotyping are making it easier for breeders to identify these sequence variants and select animals, or plants, with improved qualities and greater resilience to disease or environmental stress. This is key to increasing food security.
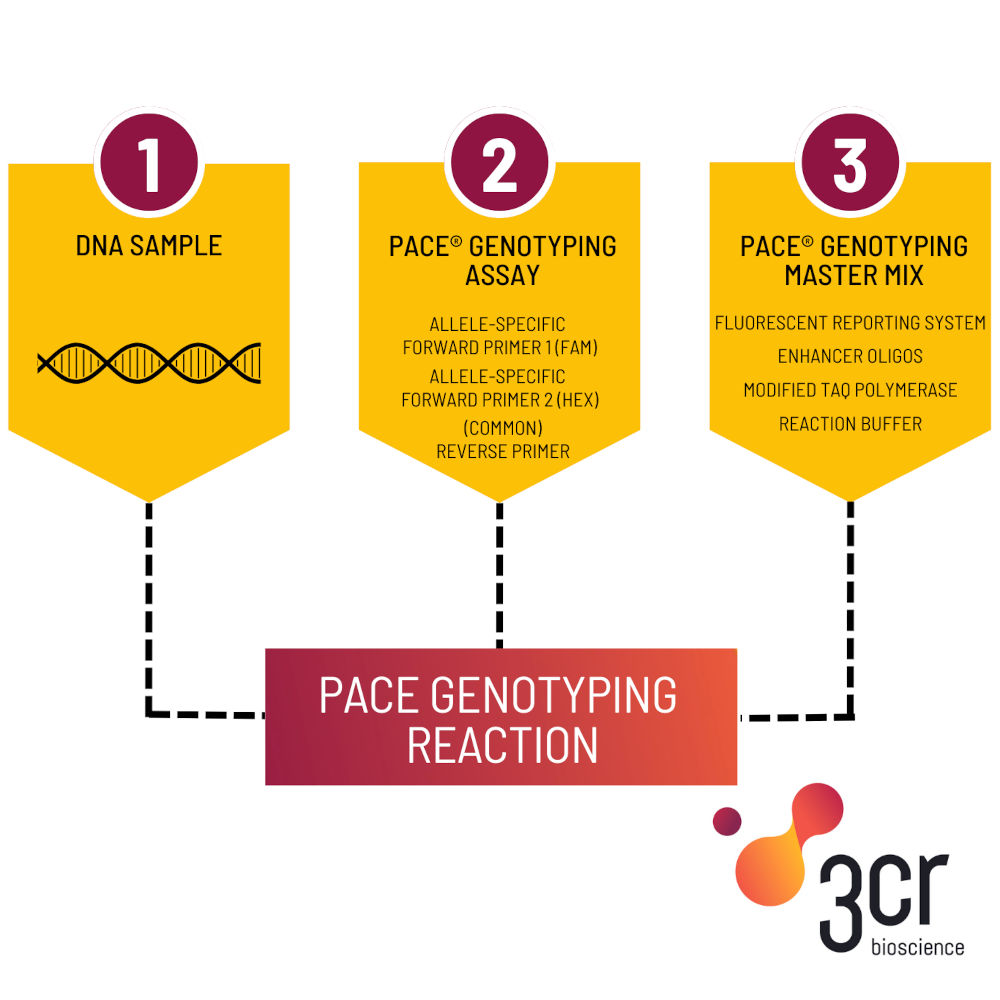
3CR Bioscience is a leader in PCR genotyping technology and has developed a patented range of reagents (PACE®) and tools that can accelerate many applications in plant and animal breeding. This includes marker-assisted breeding, pathogen detection, and gene editing, which can significantly reduce both time and costs for the breeder.


Dr Sarah Holme of 3CR Bioscience explains: “DNA sequence variants called SNPs (Single Nucleotide Variants) act as useful biomarkers for breeders.
“3CR Bioscience offers a suite of productivity tools for SNP genotyping and has developed a patented range of PCR reagents (PACE®) for replicating the desired section of DNA for analysis.
“With these tools and reagents, breeders can quickly validate markers, implement genomic selection, and conduct marker-assisted selection, thereby identifying and selecting animals with desirable traits more rapidly.”
Overcoming loss of lambs
A recent project has used PACE PCR genotyping to reduce early loss of lambs in French dairy sheep**.
High mortality rates are attributable to a variety of genetic and environmental factors. Some recessive genomic variants are known to be lethal if they are present in both the ram and the ewe.
A team at INRAE in France used PACE PCR to identify these causal variants in multiple key genes. With this knowledge, it will become possible to improve the selection of rams and improve lamb survival rates.
Sarah continues: “The speed and accuracy of PACE genotyping facilitates the rapid analysis of large numbers of animals, this aids the understanding of genetic relationships for animal health as well as evolutionary patterns, and conservation efforts.”
3CR Bioscience is talking about SNP genotyping in the Agri-TechE Innovation Hub at the Royal Norfolk Show 2024 on 26-27th June. The hub is sponsored by BBRO.
*AHDB Reducing lamb losses for higher returns: projectblue.blob.core.windows.net/media/Default/About AHDB/Reducing Lamb Losses 2020.pdf
**Searching for homozygous haplotype deficiency in Manech Tête Rousse dairy sheep revealed a nonsense variant in the MMUT gene affecting newborn lamb viability. Ben Braiek et al Genetics Selection Evolution volume 56, Article number: 16 (2024)
 Agri-TechE
Agri-TechE 




